import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import statsmodels.formula.api as smf# same link as for finches -> carnivora
df = pd.read_csv("https://raw.githubusercontent.com/roualdes/data/refs/heads/master/carnivora.csv")df.head()| Order | SuperFamily | Family | Genus | Species | FW | SW | FB | SB | LS | GL | BW | WA | AI | LY | AM | IB | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Carnivora | Caniformia | Canidae | Canis | Canis lupus | 31.1 | 33.1 | 130.0 | 132.3 | 5.5 | 63.0 | 425.0 | 35.0 | NaN | 177.0 | 913 | 12 |
| 1 | Carnivora | Caniformia | Canidae | Canis | Canis latrans | 9.7 | 10.6 | 84.5 | 88.3 | 6.2 | 61.5 | 225.0 | 98.0 | NaN | NaN | 365 | 12 |
| 2 | Carnivora | Caniformia | Canidae | Canis | Canis adustus | 10.6 | 11.3 | 53.5 | 51.8 | 4.3 | 63.3 | NaN | NaN | NaN | 127.0 | NaN | NaN |
| 3 | Carnivora | Caniformia | Canidae | Canis | Canis mesomelas | 7.2 | 7.7 | 52.0 | 56.8 | 3.8 | 60.0 | NaN | 61.0 | 270.0 | NaN | 392 | NaN |
| 4 | Carnivora | Caniformia | Canidae | Lycaon | Lycaon pictus | 22.2 | 22.0 | 128.0 | 129.0 | 8.8 | 70.5 | 365.0 | 77.0 | 390.0 | 132.0 | t 132 | 13 |
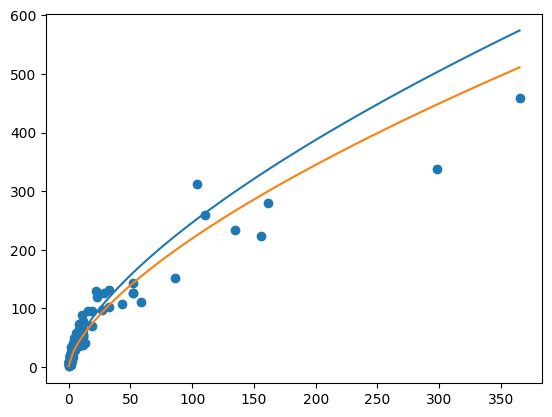
plt.scatter(df["SW"], df["SB"])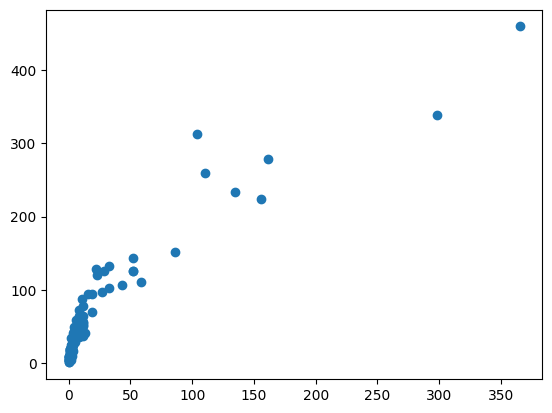
model = smf.ols("SB ~ SW + I(SW* SW)", data = df)
fit = model.fit()fit.summary()| Dep. Variable: | SB | R-squared: | 0.915 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.913 |
| Method: | Least Squares | F-statistic: | 585.6 |
| Date: | Thu, 20 Nov 2025 | Prob (F-statistic): | 4.93e-59 |
| Time: | 08:58:06 | Log-Likelihood: | -503.95 |
| No. Observations: | 112 | AIC: | 1014. |
| Df Residuals: | 109 | BIC: | 1022. |
| Df Model: | 2 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 21.3680 | 2.423 | 8.818 | 0.000 | 16.565 | 26.171 |
| SW | 2.2409 | 0.110 | 20.355 | 0.000 | 2.023 | 2.459 |
| I(SW * SW) | -0.0032 | 0.000 | -8.822 | 0.000 | -0.004 | -0.003 |
| Omnibus: | 26.840 | Durbin-Watson: | 1.275 |
|---|---|---|---|
| Prob(Omnibus): | 0.000 | Jarque-Bera (JB): | 70.392 |
| Skew: | 0.848 | Prob(JB): | 5.18e-16 |
| Kurtosis: | 6.494 | Cond. No. | 1.83e+04 |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
[2] The condition number is large, 1.83e+04. This might indicate that there are
strong multicollinearity or other numerical problems.
mn, mx = np.min(df["SW"]), np.max(df["SW"])
ndf = pd.DataFrame.from_dict({
"SW": np.linspace(mn, mx, 101)
})
ndf["yhat"] = fit.predict(ndf)
plt.scatter(df["SW"], df["SB"])
plt.plot(ndf["SW"], ndf["yhat"], color = "black")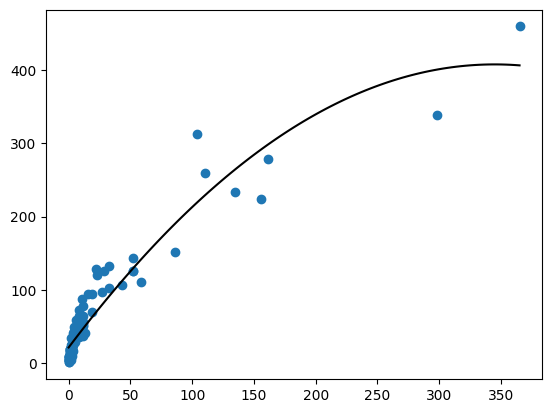
plt.scatter(np.log10(df["SW"]), np.log10(df["SB"]))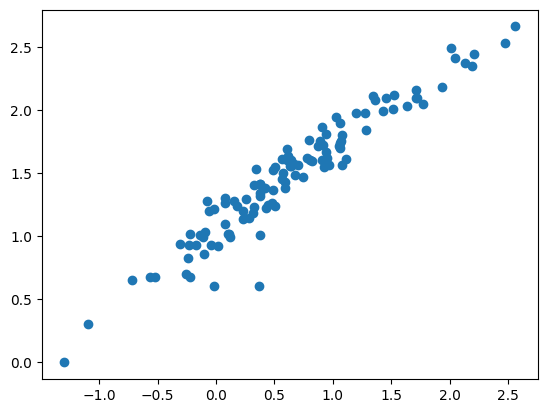
model = smf.ols("np.log10(SB) ~ np.log10(SW)", data = df)
fit = model.fit()yhat = fit.predict(df)
plt.scatter(df["SW"], df["SB"])
plt.plot(df["SW"], yhat)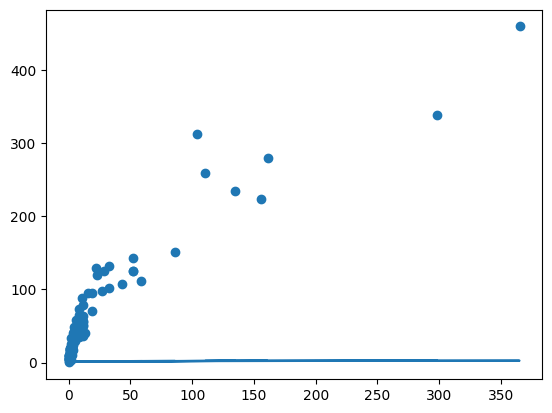
mn, mx = np.min(df["SW"]), np.max(df["SW"])
ndf = pd.DataFrame.from_dict({
"SW": np.linspace(mn, mx, 101)
})
ndf["yhat"] = fit.predict(ndf)
plt.scatter(df["SW"], df["SB"])
plt.plot(ndf["SW"], 10 ** ndf["yhat"], color = "black")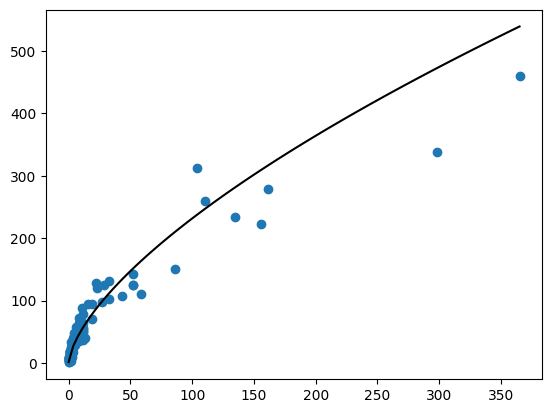
# instantiate and fit model here: log10 both x, y, and
# unique "intercepts" by SuperFamily
mn, mx = np.min(df["SW"]), np.max(df["SW"])
x = np.linspace(mn, mx, 101)
ndf = pd.DataFrame.from_dict({
"SuperFamily": np.repeat(["Caniformia", "Feliformia"], 101),
"SW": np.tile(x, 2)
})
ndf["yhat"] = fit.predict(ndf)
# make plot here: one line for each SuperFamily