import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import statsmodels.api as sm
import statsmodels.formula.api as smf
import patsy as pt Linear Regression
hospital = pd.read_csv("https://raw.githubusercontent.com/roualdes/data/refs/heads/master/hospital.csv")plt.scatter(hospital["stay"], hospital["infection_risk"])
plt.xlabel("Stay (days)")
plt.ylabel("Infection risk");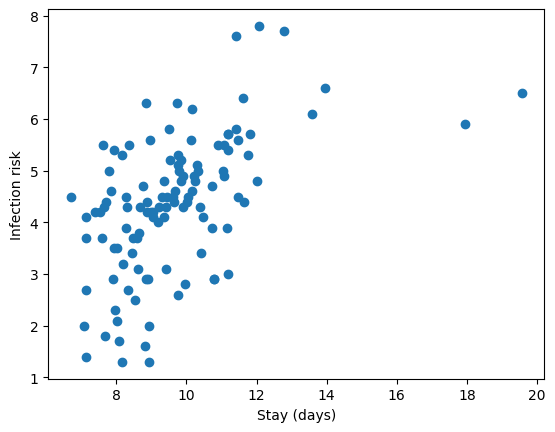
fit = smf.ols("infection_risk ~ nurses * stay", data = hospital).fit()
fit.summary()| Dep. Variable: | infection_risk | R-squared: | 0.349 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.331 |
| Method: | Least Squares | F-statistic: | 19.48 |
| Date: | Tue, 02 Dec 2025 | Prob (F-statistic): | 3.48e-10 |
| Time: | 22:24:29 | Log-Likelihood: | -168.73 |
| No. Observations: | 113 | AIC: | 345.5 |
| Df Residuals: | 109 | BIC: | 356.4 |
| Df Model: | 3 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | -0.3488 | 0.989 | -0.353 | 0.725 | -2.308 | 1.611 |
| nurses | 0.0089 | 0.004 | 1.993 | 0.049 | 5.08e-05 | 0.018 |
| stay | 0.4498 | 0.106 | 4.253 | 0.000 | 0.240 | 0.659 |
| nurses:stay | -0.0007 | 0.000 | -1.499 | 0.137 | -0.002 | 0.000 |
| Omnibus: | 0.593 | Durbin-Watson: | 1.878 |
|---|---|---|---|
| Prob(Omnibus): | 0.744 | Jarque-Bera (JB): | 0.716 |
| Skew: | 0.153 | Prob(JB): | 0.699 |
| Kurtosis: | 2.758 | Cond. No. | 2.27e+04 |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
[2] The condition number is large, 2.27e+04. This might indicate that there are
strong multicollinearity or other numerical problems.
ndf = pd.DataFrame.from_dict({"stay": [20, 20], "nurses": [100, 101]})
np.diff(fit.predict(ndf))array([-0.00447639])For each extra nurse, when stay is equal to 20, we expect infection risk to decrease by 0.004.
ndf = pd.DataFrame.from_dict({"stay": [10, 10], "nurses": [100, 101]})
np.diff(fit.predict(ndf))array([0.00221305])For each extra nurse, when stay is equal to 10, we expect infection risk to increase by 0.002.
Logistic Regression
# change hospital -> possum
possum = pd.read_csv("https://raw.githubusercontent.com/roualdes/data/refs/heads/master/possum.csv")possum["vic"] = (possum["pop"].astype("string") == "vic").astype(np.float64)rng = np.random.default_rng()
N = np.shape(possum)[0]
plt.scatter(possum["tailL"] + rng.uniform(size = N) * 0.25, possum["vic"],
alpha = 0.25);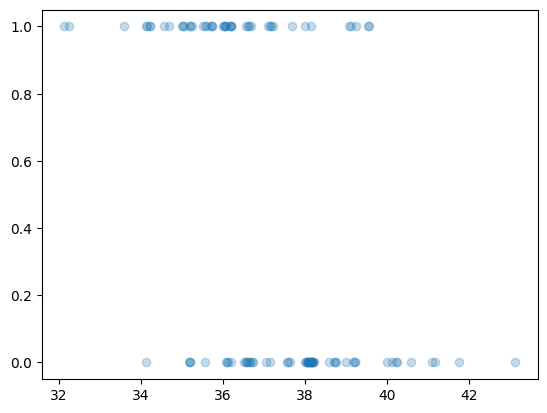
fit = smf.glm("vic ~ age * tailL", data = possum,
family = sm.families.Binomial()).fit()fit.summary()| Dep. Variable: | vic | No. Observations: | 102 |
|---|---|---|---|
| Model: | GLM | Df Residuals: | 98 |
| Model Family: | Binomial | Df Model: | 3 |
| Link Function: | Logit | Scale: | 1.0000 |
| Method: | IRLS | Log-Likelihood: | -53.011 |
| Date: | Tue, 02 Dec 2025 | Deviance: | 106.02 |
| Time: | 22:24:29 | Pearson chi2: | 113. |
| No. Iterations: | 5 | Pseudo R-squ. (CS): | 0.2796 |
| Covariance Type: | nonrobust |
| coef | std err | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 49.3407 | 15.472 | 3.189 | 0.001 | 19.016 | 79.665 |
| age | -5.5348 | 3.032 | -1.825 | 0.068 | -11.478 | 0.409 |
| tailL | -1.3680 | 0.422 | -3.239 | 0.001 | -2.196 | -0.540 |
| age:tailL | 0.1554 | 0.082 | 1.887 | 0.059 | -0.006 | 0.317 |
x = np.linspace(32, 44, 101)
m = np.mean(possum["age"])
ndf = pd.DataFrame.from_dict({"tailL": x, "age": np.full(101, m)})
yhat = fit.predict(ndf)plt.scatter(possum["tailL"] + rng.uniform(size = N) * 0.25, possum["vic"],
alpha = 0.25);
plt.plot(x, yhat);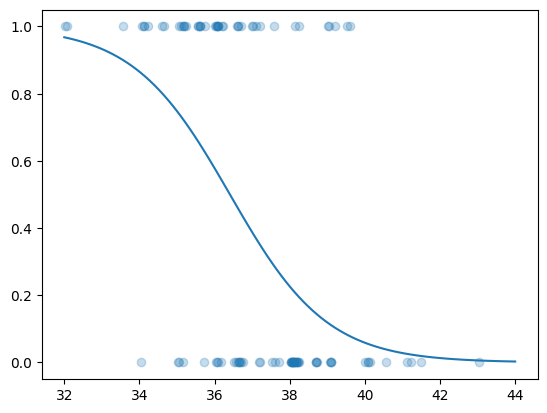
ndf = pd.DataFrame.from_dict({"tailL": [37, 38], "age": [m, m]})
np.diff(fit.predict(ndf))array([-0.16145695])