---
toc: false
format: html
---
# MATH 315 Practice Exam 02
```{r}
#| echo: false
#| output: false
library(dplyr)
library(ggplot2)
```
1. Describe the Central Limit Theorem and also include why it's appropriate to use a symmetric confidence interval.
2. Consider the variable `LS` (litter size) from the carnivora dataset. The output of a t-test is below. Answer the following question based on this output.
```{r}
carnivora <- read.csv("https://raw.githubusercontent.com/roualdes/data/refs/heads/master/carnivora.csv")
t.test(carnivora$LS, mu = 2, conf.level = 0.9)
```
a. How many observations were measured / what is the sample size?
b. Interpret, in context of these data, the confidence interval.
c. Did this confidence interval capture the true population
parameter it is targeting?
d. If we had instead more observations in the sample, would the confidence interval
be narrower or wider? Explain why, using concepts and keywords from
this class.
e. If we instead made a 98% confidence interval, would the
confidence interval be narrower or wider? Explain why, using concepts
and keywords from this class.
3. Consider again the carnivora dataset, where GL represents gestation length in days and WA represents weaning age in days. The output of a t-test is below.
Answer the following questions based on this output.
```{r}
carnivora$diff <- carnivora$GL - carnivora$WA
t.test(carnivora$diff, conf.level = 0.99)
```
a. What type of hypothesis test is this, what is the proper name?
b. Write the corresponding null and alternative hypotheses using proper statistical symbols.
c. What is the value for the level of significance?
d. Write a statistical conclusion using the p-value.
e. Write an interpretation of the conclusion from d. with minimal
statistical jargon.
f. Explain why the confidence interval supports the same conclusion
as the p-value.
4. Consider again the carnivora dataset. The output of a t-test is below.
Answer the following questions based on this output.
```{r}
t.test(LS ~ SuperFamily, data = carnivora, conf.level = 0.9)
```
a. What type of hypothesis test is this, what is the proper name?
b. Write the corresponding null and alternative hypotheses using
proper statistical symbols.
c. What does this output tell you about these data? Write a
conclusion with little to no statistical jargon, citing the
hypothesis test or the confidence interval to support your claim.
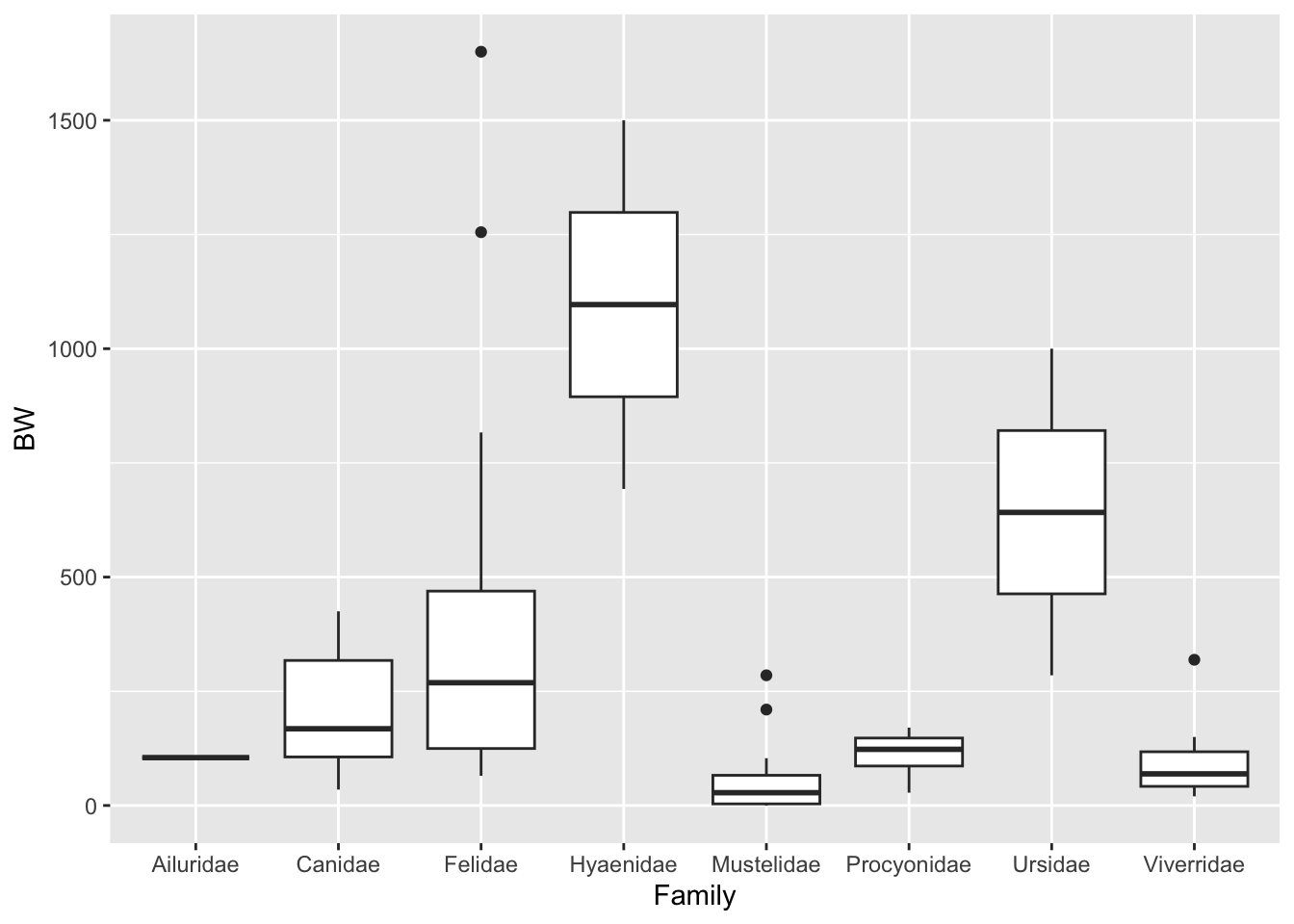
5. Consider again the carnivora dataset, where BW represents birth weight in grams. Based on the following code, answer the questions below.
```{r}
ggplot(carnivora, aes(Family, BW)) + geom_boxplot()
fit <- lm(BW ~ Family, data = carnivora)
anova(fit)
TukeyHSD(aov(fit))
```
a. Is the group degrees of freedom correct? Explain.
b. What is the appropriate statistical conclusion from this test?
c. Write an interpretation of the conclusion with minimal statistical jargon.
d. Explain why it is appropriate to use Tukey's HSD here.
e. What does Tukey's HSD tell you about these possums?