MATH 314 Exam 02
Exam 02 will take place during the week of May 5 - 8,
during our regular class hours and in our regular
classroom. Please schedule your preferred time from this Google spreadsheet.
You forfeit your right to take Exam 02 by moving someone
else's name on this spreadsheeet.
For Exam 02, I need to give you some starter code. This repository will contain the appropriate starter code for your Exam 02. https://classroom.github.com/a/R3YDXhiI
Please clone now this repository to your
local machine, so that you can git pull your
starter code right before taking Exam 02.
Only these imports are allowed on Exam 02.
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import patsy as pt
from scipy.optimize import minimize
Here's some functions you can use during Exam 02.
def lm(theta, X):
return X @ theta
def mse(y, yhat):
d = y - yhat
return np.mean(d * d)
def normal_ll(theta, data):
y = data['y']
X = data['X']
yhat = lm(theta, X)
return mse(y, yhat)
def windows(N, K):
w = N // K
ws = []
for k in range(K):
start = k * w
end = (k + 1) * w
if k == K - 1:
end = N
ws.append((start, end))
return ws - In general, we should prefer models that predict better on
data the model has never seen / been trained on. A common way to
measure such prediction accuracy, when one only has one dataset, is to
perform -fold cross validation. If ,
then we will randomly split the dataset up into (roughly
equal) folds. Then, cycling across all folds, train the model on
out of folds, and
measure prediction accuracy on the last (test) fold.
Whichever model has lower average (across all test folds)
prediction error is considered better.
Use the hospital dataset and -fold cross validation to determine which model is a better predictor ofinfection_risk. Consider a model that usesstayversus a model that usesstayandstay ** 2. Use . Here's some starter code:... rng = np.random.default_rng() N = ... idx = rng.permutation(np.arange(N)) K = 5 mses = np.zeros(K) mse2s = np.zeros(K) ws = windows(N, K) for k in range(K): b, e = ws[k] jdx = idx[b:e] # test indices ndx = np.setdiff1d(idx, jdx) # train indices ... (np.mean(mses), np.mean(mse2s)) - Write a class named
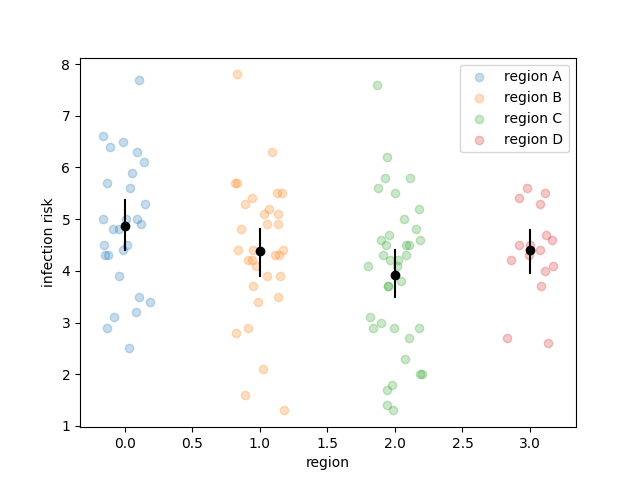
LinearRegressionthat implements the following APIlr = LinearRegression('infection_risk ~ stay', df) lr.fit() betas = lr.bootstrap() plt.scatter(betas[:, 0], betas[:, 1]);Your code should work for the example above. Further, for full credit, I will test your code during the exam with a simple change to the code above. Here's the structure of the class I recommendclass LinearRegression(): def __init__(self, ptsystr, df): ... def _lm(self, theta, X): ... def _mse(self, y, yhat): ... def fit(self, y = None, X = None): ... def bootstrap(self, R = 1_000): ...During Exam 02, I'll ask you to recreate just one of the methods from the above class, and I'll supply the code for the other methods. - Recreate the following plot. The black lines are
bootstrapped confidence intervals and
the black dots are medians ( quantiles)
which are also calculated from the bootstrap resampled
coefficients.
There are three distinct steps to recreate this plot:
- Bootstrap the appropriate model
- Calculate confidence intervals from the estimates of the bootstrapped coefficients
- make the plot -- hint: learn that you can loop over the output of
df.groupby(...)
- The Rayleigh distribution has density function
for .
Use the following data to find the
maximum likelihood estimator of and
to produce a confidence
interval using the bootstrap method.
[2.55949718, 4.52940383, 3.97518473, 3.38191934, 1.40503558, 1.52411053, 6.91676848, 1.45665845, 2.3602429 , 1.60873959, 3.94374552, 6.22249149, 1.89894113, 5.20759437, 7.24625107, 2.80471966, 4.81060617, 3.08315807, 4.72002917, 1.2751267 , 6.66914935] - Show, empirically (with code), that the procedure to create a confidence
interval does indeed capture the expectation value in
roughly of confidence intervals.
Your code should repeat the following times
- generate data from the exponential distribution with rate parameter
- calculate confidence interval (not bootstrap)
- determine if the confidence interval captures the expectation, call this value
c - increment a running mean with
cusing the method defined in Homework 03